Looking at changes in vegetative health at Glacier National Park, 2015-2020¶
Glacier National Park is warming at nearly two times the global average [1] threatening the very glaciers after which the park is named [2]. The plants and animals that call the park home are also being impacted by warming, including the potato-sized, furry, North American pika (Ochotona princeps) [3]. Additional stressors to the forest ecosystems inside Glacier National Park include various rusts, worms, and beetles [4], as well as fire [5].
 Grinnell Glacier; Glacier National Park; Image Credit: 1910-Elrod photo, U of M Collection and 2016-McKeon photo, USGS
Grinnell Glacier; Glacier National Park; Image Credit: 1910-Elrod photo, U of M Collection and 2016-McKeon photo, USGS
References¶
- [1] https://www.nps.gov/glac/learn/nature/climate-change.htm
- [2] https://www.usgs.gov/centers/norock/science/repeat-photography-project
- [3] https://www.nps.gov/articles/pikas-brief.htm
- [4] https://www.nps.gov/articles/forest-health-brief.htm
- [5] https://www.nps.gov/glac/learn/nature/fire-history.htm
An open Environmental Data Science Workflow¶
Below is an open EDS workflow that analyzes NDVI data from MODIS accessed using the NASA AppEEARS API. Through this analysis we calcuate the difference in NDVI between two 3-year time periods (2015-2017 and 2018-2020), along with looking at differences in NDVI inside and outside the park from 2015-2020. This analysis resulted in the two following plots that are summarized with the brief headline and description. The full analysis is also included, as well as the code written to create the plots.
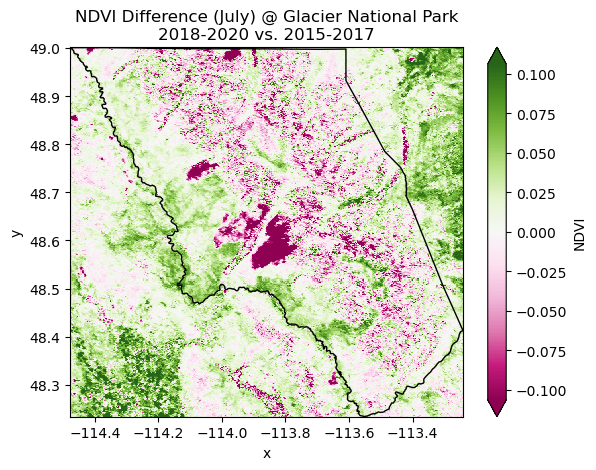
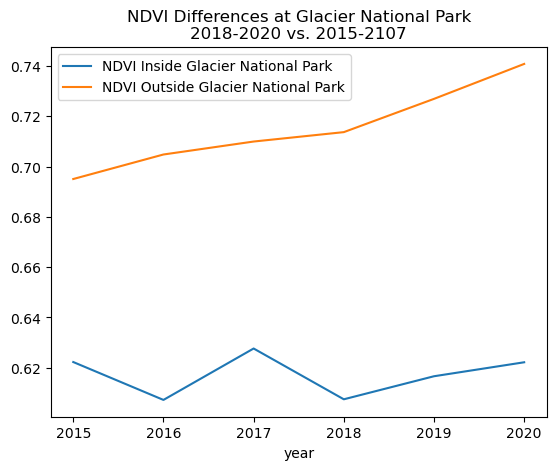
Glacier National Park is greening up ... sort of?¶
Comparing normalized difference vegetative index (NDVI) data from 2015-2020 at Glacier National Park reveals that the area inside and outside the park is getting greener over time. The data from this time period also reveal a number of burn scars that are likely from the Howe Ridge (2018) and Sprague (2017) fires near Lake McDonald [5]. There is also widespread decrease in vegetative health throughout the park, potentially due to beetle kill or other forest pathogens. More data would be needed to confirm this hypothesis.
 Boulder Glacier; Glacier National Park; Image Credit: Morton Elrod, 1910, U of M Collection, Fagre/Pederson, 2007, USGS
Boulder Glacier; Glacier National Park; Image Credit: Morton Elrod, 1910, U of M Collection, Fagre/Pederson, 2007, USGS
Complete workflow¶
Below is the entire workflow needed to complete this analysis including code to access the NASA AppEEARS API an creating the plots shown above.
# Import python libraries
import getpass
import json
import os
import pathlib
from glob import glob
import pandas as pd
import geopandas as gpd
import matplotlib.pyplot as plt
import earthpy.appeears as eaapp
import hvplot.pandas
import hvplot.xarray
import rioxarray as rxr
import xarray as xr
Creating a boundary for Glacier National Park¶
# Read in all National Park Boundaries (US)
nps_boundary = gpd.read_file("http://gstore.unm.edu/apps/rgisarchive/"
"datasets/7bbe8af5-029b-4adf-b06c-134f0dd57226/"
"nps_boundary.original.zip")
nps_boundary
| GIS_LOC_ID | UNIT_CODE | GROUP_CODE | UNIT_NAME | UNIT_TYPE | META_MIDF | LANDS_CODE | DATE_EDIT | GIS_NOTES | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | None | UPDE | None | Upper Delaware | Scenic and Recreational River | None | None | 2006-06-06 | POC for this update: leslie_morlock@nps.gov -... | POLYGON ((-74.92112 41.49030, -74.91918 41.491... |
| 1 | None | BAND | None | Bandelier | National Monument | None | None | None | Park Created Boundary - contact Kay Beeley | MULTIPOLYGON (((-106.19585 35.85378, -106.2214... |
| 2 | None | ACAD | None | Acadia | National Park | None | None | None | Shift 0.03 mi. | MULTIPOLYGON (((-68.22595 44.39584, -68.22766 ... |
| 3 | None | ROCR | None | Achbold Parkway | Park | None | None | None | None | MULTIPOLYGON (((-77.08430 38.91673, -77.08482 ... |
| 4 | None | ADAM | None | Adams | National Historical Park | None | None | None | Shift 0.03 mi. | POLYGON ((-71.00948 42.25625, -71.01133 42.255... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 505 | None | CLBA | None | Clara Barton | National Historic Site | None | None | 2008-11-17 | Lands - http://landsnet.nps.gov/tractsnet/docu... | POLYGON ((-77.14072 38.96699, -77.14075 38.967... |
| 506 | None | FOWA | None | Fort Washington Park | Park | None | None | 2008-11-17 | Lands - http://landsnet.nps.gov/tractsnet/docu... | POLYGON ((-77.02299 38.72054, -77.02032 38.718... |
| 507 | None | KICA | None | Kings Canyon | National Park | None | None | 2003-11-07 | Lands | MULTIPOLYGON (((-118.77919 37.23609, -118.7791... |
| 508 | None | SEQU | None | Sequoia | National Park | None | None | 2004-01-09 | Lands | POLYGON ((-118.70637 36.66475, -118.70636 36.6... |
| 509 | None | CABR | None | Cabrillo | National Monument | None | None | 2008-08-28 | Lands - http://landsnet.nps.gov/tractsnet/docu... | POLYGON ((-117.23862 32.66936, -117.24036 32.6... |
510 rows × 10 columns
# Select out Glacier National Park
glacier = nps_boundary[nps_boundary["UNIT_NAME"] == "Glacier"]
glacier
| GIS_LOC_ID | UNIT_CODE | GROUP_CODE | UNIT_NAME | UNIT_TYPE | META_MIDF | LANDS_CODE | DATE_EDIT | GIS_NOTES | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|
| 361 | None | GLAC | None | Glacier | National Park | None | None | 2006-03-09 | POC for this update: richard_menicke@nps.gov ... | POLYGON ((-114.47552 49.00091, -114.46101 49.0... |
# Plot the boundary to inspect
glacier.explore()
# Create a path to the data
glacier_dir = os.path.join(pathlib.Path.home(), 'glacier-data')
# Make the data directory
os.makedirs(glacier_dir, exist_ok=True)
glacier_dir
'/home/jovyan/glacier-data'
Downloading and preparing the data¶
# Initialize AppeearsDownloader for MODIS NDVI data
ndvi_downloader = eaapp.AppeearsDownloader(
download_key='glacier-ndvi',
ea_dir=glacier_dir,
product='MOD13Q1.061',
layer='_250m_16_days_NDVI',
start_date="07-01",
end_date="07-31",
recurring=True,
year_range=[2015, 2020],
polygon=glacier
)
ndvi_downloader.download_files(cache=True)
# Get a list of NDVI tif file paths
glacier_paths = sorted(glob(os.path.join(glacier_dir, 'glacier-ndvi', '*', '*NDVI*.tif')))
len(glacier_paths)
18
scale_factor = 10000
doy_start = -19
doy_end = -12
# Build data list of data arrays
glacier_das = []
for glacier_path in glacier_paths:
# Get date from file name
doy = glacier_path[doy_start:doy_end]
date = pd.to_datetime(doy, format='%Y%j')
# Open dataset
da = rxr.open_rasterio(glacier_path, masked=True).squeeze()
# Add date dimension and clean up metadata
da = da.assign_coords({'date': date})
da = da.expand_dims({'date': 1})
da.name = 'NDVI'
# Multiple by scale factor
da = da / scale_factor
# Prepare for concatenation
glacier_das.append(da)
len(glacier_das)
18
# Combine into single array by date
glacier_da = xr.combine_by_coords(glacier_das, coords=['date'])
glacier_da
<xarray.Dataset>
Dimensions: (x: 593, y: 369, date: 18)
Coordinates:
band int64 1
* x (x) float64 -114.5 -114.5 -114.5 ... -113.2 -113.2 -113.2
* y (y) float64 49.0 49.0 49.0 48.99 ... 48.24 48.24 48.24 48.23
spatial_ref int64 0
* date (date) datetime64[ns] 2015-06-26 2015-07-12 ... 2020-07-27
Data variables:
NDVI (date, y, x) float32 0.5447 0.5447 0.6585 ... 0.7097 0.7097# Compute the difference in NDVI before and 2017/2018
glacier_diff = (
glacier_da
.sel(date=slice('2018', '2020'))
.mean('date')
.NDVI
- glacier_da
.sel(date=slice('2015', '2017'))
.mean('date')
.NDVI
)
glacier_diff
<xarray.DataArray 'NDVI' (y: 369, x: 593)>
array([[ 0.00964445, 0.00964445, -0.00370002, ..., 0.07951105,
0.09175545, 0.09175545],
[ 0.03782219, 0.03242224, 0.01788896, ..., 0.09422225,
0.07023335, 0.0790112 ],
[ 0.03799999, 0.03799999, 0.04133326, ..., 0.06477785,
0.06571102, 0.06571102],
...,
[ 0.00105554, 0.00936663, 0.00936663, ..., -0.00990003,
-0.02216661, -0.02216661],
[ 0.01837778, 0.01837778, 0.00757778, ..., -0.03024441,
-0.03024441, -0.03652221],
[ 0.01888889, 0.01841098, 0.01841098, ..., -0.04922229,
-0.01131105, -0.01131105]], dtype=float32)
Coordinates:
band int64 1
* x (x) float64 -114.5 -114.5 -114.5 ... -113.2 -113.2 -113.2
* y (y) float64 49.0 49.0 49.0 48.99 ... 48.24 48.24 48.24 48.23
spatial_ref int64 0Plotting the NDVI difference spatially¶
NOTE: Remember to try the .hvplot approach now that the environment is rebuilt?
# Plot the difference
fig, ax = plt.subplots()
glacier_diff.plot(x='x', y='y', cmap='PiYG', robust=True, ax=ax)
glacier.plot(edgecolor='black', color='none', ax=ax)
plt.title('NDVI Difference (July) @ Glacier National Park\n2018-2020 vs. 2015-2017')
plt.show()
Looking at how NDVI changes with time¶
glacier_out_gdf = (
gpd.GeoDataFrame(geometry=glacier.envelope)
.overlay(glacier, how='difference'))
# Clip data to both inside and outside the boundary
ndvi_glacier_da = glacier_da.rio.clip(glacier.geometry, from_disk=True)
ndvi_glacier_out_da = glacier_da.rio.clip(glacier_out_gdf.geometry, from_disk=True)
# Compute mean annual July NDVI
jul_ndvi_glacier_df = (
ndvi_glacier_da
.groupby(ndvi_glacier_da.date.dt.year)
.mean(...)
.NDVI.to_dataframe())
jul_ndvi_glacier_out_df = (
ndvi_glacier_out_da
.groupby(ndvi_glacier_out_da.date.dt.year)
.mean(...)
.NDVI.to_dataframe())
# Plot inside and outside the reservation
glacier_ndvi_df = (
jul_ndvi_glacier_df[['NDVI']]
.join(
jul_ndvi_glacier_out_df[['NDVI']],
lsuffix=' Inside Glacier National Park', rsuffix=' Outside Glacier National Park')
)
# Plot difference inside and outside the reservation
glacier_ndvi_df.plot(
title='NDVI Differences at Glacier National Park\n2018-2020 vs. 2015-2107')
<Axes: title={'center': 'NDVI Differences at Glacier National Park\n2018-2020 vs. 2015-2107'}, xlabel='year'>
Create a .html (or markdown) copy of your work¶
%%capture
%%bash
jupyter nbconvert *.ipynb --to html